COVID-19 variants can't hide from Variabel
Details about variants hiding in the deluge of genetic SARS-CoV-2 sequences would be good to know, if only researchers could get to them.
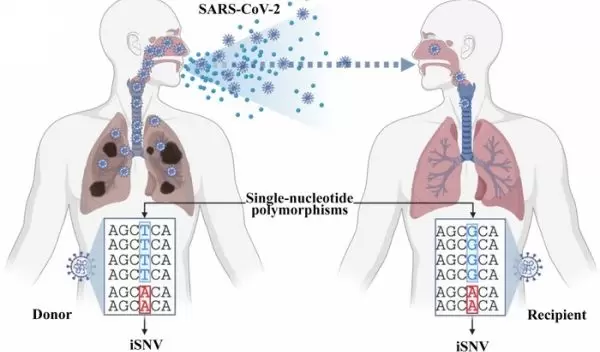
A U.S. National Science Foundation-funded program developed at Rice University will make that possible, at least for "intrahost variants" -- variants that appear in genome data from the same COVID-19-positive person.
"This grant enables analyses of viral genomic datasets, especially important during a pandemic," said Deepankar Medhi, a program director in NSF's Division of Computer and Network Systems. "NSF's long-term investment in computational resources is important for scientists working on research projects like this one."
A team led by computer scientists Todd Treangen and Yunxi Liu developed Variabel, a program which accurately identifies "low-frequency variants" of the SARS-CoV-2 virus, which causes COVID-19. Finding these clues could be the key to identifying potentially devastating variants before they have a chance to spread, according to Treangen.
"Variabel enables the use of affordable nanopore sequencing technology for the identification of within-host variation after viral infection," said Treangen. The lab had similar success in testing Variabel on sequence data from patients infected with Ebola and norovirus. The open-source program is detailed in Nature Communications.
The researchers claim the key to Variabel is its ability to distinguish true variants from sequencing errors. To validate Variabel, they compared data taken over time from single positive patients, as well as sequences from cross-patient datasets. Over time, a single patient can host as many as a billion copies of a virus.
By comparing results before and after applying Variabel to the data, the researchers found the program was able to correct the majority of sequencing errors.
"Variabel opens the door to portable, affordable and rapid characterization of within-host variation, which could aid in the discovery of future mutations specific to variants of concern," said Treangen, whose lab hosted a symposium to discuss scientific advances spurred by the pandemic. The virtual symposium may be viewed online at http://www.youtube.com/watch?v=YaNm7QBmxD8.


