Molecular Self-Assembly Technique May Mimic How Cells Assemble Themselves
Researchers from the University of Pennsylvania and the University of Sheffield have created tree-like molecules that assemble themselves into precisely structured building blocks of a quarter-million atoms. Such building blocks may be precursors to designing nanostructures for molecular electronics or photonics materials, which "steer" light in the same way computer chips steer electrons.
Virgil Percec, the P. Roy Vagelos Chair and Professor of Chemistry at the University of Pennsylvania, and his colleagues also provided chemists with pointers for designing variations of the tree-like molecules to form even larger-scale structures. The work is funded by NSF and the Engineering and Physical Sciences Research Council in the United Kingdom.
"Percec and his collaborators have developed a model that may mimic what happens in cell self-assembly," said NSF program officer Andrew Lovinger. "This is the first time where you get large-scale supramolecular structures to assemble themselves into such exceptionally large and complex structures."
The goal of photonics is to control light the way electronics control and use electrons. A working photonics crystal would have to be approximately as large as the light's wavelength -- on the order of hundreds or thousands of nanometers -- yet precisely structured to have predictable and reproducible interactions with the light. The techniques developed by Percec and colleagues may help chemists design self-assembling materials that approach photonics size.
"Photonics crystals require repeating units whose size is in the range of the wavelength of light," Percec said. "So far, we're the only ones who can design with the precision of atoms but at a nanometer scale. This sort of precision and behavior is previously unknown in organic chemistry."
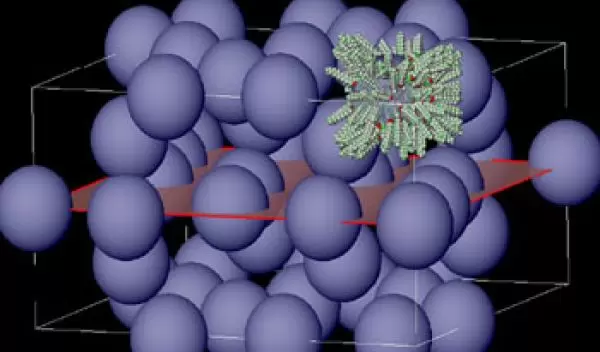
The researchers start with tree-like organic molecules, called dendrons, each of which is roughly cone-shaped. Twelve of the dendrons assemble themselves into 8,500-atom spheres. Once assembled, the spheres become a "liquid crystal," a material that flows like a liquid but has some properties of a crystalline solid. Liquid crystals are commonly found in flat-panel computer screens and many other devices.
In the right conditions, liquid crystal molecules "pack" themselves into very regular, repeating patterns, called lattices. A common lattice structure resembles neatly stacked layers of golf balls in a box. However, instead of packing into common lattices, the spheres created by Percec's team arrange themselves into much more complex formations.
"We created extremely large objects that pack into the most complex lattices rather than the simplest ones that everyone expected," Percec said. "They have lattices that we haven't seen before with organic molecules. They behave like heavy atoms, with a hard core and a soft outer part -- like the electron clouds surrounding metals such as uranium."
Because they are constructed from dendrons, the spheres aren't solid, but instead have a brush-like surface composed of the dendrons' "branches." The brush-like surface allows the spheres to deform slightly and fill space more like soap bubbles than like golf balls. However, the spheres are firm enough to create repetitive lattices.
In this case, the repetitive "building block," or unit cell, comprises 30 spheres -- more than 250,000 atoms -- in a rectangular volume nearly 20 nanometers by 10 nanometers. For comparison, the rhinoviruses responsible for many human colds have diameters of about 25 nanometers. The researcher's work, published in the journal Science in 2003, provides pointers that may allow chemists to make even larger spheres that will pack into more complex lattices that are large enough to scatter light.
A year later, Percec and colleagues built upon this work to create the first artificial analogs of nature's molecular "pores"--the tiny, hollow channels that perform a multitude of essential tasks in living cells. Writing in the journal Nature, Penn chemist Virgil Percec and his colleagues note that natural pores are used by the cell to transport certain molecules across the cell membrane, as well as to generate chemical energy, guide the shape of newly-made proteins, and even puncture holes in the cell walls of hostile bacteria.
To create the new artificial pores, Percec developed a series of small, protein-like molecules that assemble themselves into molecular channels spontaneously. Potential applications range from the extraction of fresh water from seawater to an entirely new class of antibiotics.
-- David Hart


