News Release 11-197
On-line Gamers Succeed Where Scientists Fail, Opening Door to New AIDS Drug Design
Molecular structure of retrovirus enzyme solved through "Foldit"
The "unsolved monkey virus protein" Foldit puzzle, a tool used by online gamers.
September 19, 2011
This material is available primarily for archival purposes. Telephone numbers or other contact information may be out of date; please see current contact information at media contacts.
Online gamers have solved the structure of a retrovirus enzyme whose configuration had stymied scientists.
The players were adept at a computer game, Foldit, that allows players to collaborate and compete in predicting the structure of protein molecules.
After scientists repeatedly failed to piece together the structure of a protein-cutting enzyme from an AIDS-like virus, they called in the Foldit players.
The scientists challenged the gamers to produce an accurate model of the enzyme.
This class of enzymes, called retroviral proteases, has a critical role in how the AIDS virus matures and proliferates.
Intensive research is underway to try to find anti-AIDS drugs that can block these enzymes, but efforts were hampered by not knowing exactly what the retroviral protease molecule looked like.
"We wanted to see if human intuition could succeed where automated methods had failed," said biochemist Firas Khatib of the University of Washington (UW).
Remarkably, the gamers generated models good enough for the researchers to refine and determine the enzyme's structure within a few days.
Equally amazing, surfaces on the molecule stood out as likely targets for drugs to de-active the enzyme.
"These features provide exciting opportunities for the design of retroviral drugs, including AIDS drugs," write Khatib and co-authors of a paper appearing yesterday in the journal Nature Structural & Molecular Biology.
The scientists and gamers are all listed as co-authors.
"Online gamers have solved a longstanding scientific problem, perhaps leading the way to new anti-viral drugs," said Carter Kimsey, program director in the National Science Foundation's (NSF) Division of Biological Infrastructure, which funded the research.
"After this discovery, young people might not mind doing their online science homework," said Kimsey. "This is an innovative approach to getting humans and computer models to ‘learn from each other' in real-time."
Fold-it was created by computer scientists and biochemists at the UW Center for Game Science, and by paper co-author biochemist David Baker of UW, to engage the general public in scientific discovery.
The solution of the virus enzyme structure, the scientists said, indicates the power of online computer games to channel human intuition and of three-dimensional pattern-matching skills to solve challenging scientific problems.
With names like Foldit Contenders Group and Foldit Void Crushers Group, the gamers were excited about the task of real-world molecule-modeling problems.
The online protein folding game captivates thousands of avid players worldwide.
Players come from all walks of life. The game taps into their 3-D spatial abilities to rotate chains of amino acids in cyberspace.
New players start at the basic level, "One Small Clash," proceed to "Swing it Around," and step ahead until reaching "Rubber Band Reversal."
Direct manipulation tools, as well as assistance from a computer program called Rosetta, encourage participants to configure graphics into a workable protein model.
Teams send in their answers, and UW researchers constantly improve the design of the game and its puzzles by analyzing the players' problem-solving strategies.
Foldit is much more than a mind-boggling diversion.
As one player put it, the No. 1 reason to "crunch Rosetta at home and why you should, too, is a compelling mission: technology that could basically wipe out disease."
Figuring out the shape and misshape of proteins contributes to research on causes of and cures for cancer, Alzheimer's, immune deficiencies, and a host of other disorders, as well as to environmental work on biofuels.
Seth Cooper, a computer scientist at UW, is a co-creator of Foldit and its lead designer and developer, as well as a co-author of the paper. He studies human-computer exploration methods and the co-adaptation of games and players.
"People have spatial reasoning, something computers are not yet good at," Cooper said.
"Games provide a framework for bringing together the strengths of computers and humans. These results show that gaming, science and computation can be combined to make advances that were not possible before."
Games like Foldit are evolving. To piece together the retrovirus enzyme structure, Cooper said, Foldit used a new alignment tool for the first time to copy parts of known molecules and test their fit in an incomplete model.
"The ingenuity of game players," the paper concludes, "is a formidable force that, if properly directed, can be used to solve a wide range of scientific problems."
The other scientists involved in the project were Frank DiMaio, James Thompson and Zoran Popovic, director of the UW Center for Game Science; Macjiej Kazmierczyk, Miroslaw Gilski, Szymon Krsywda, Helena Zabranska, and Mariusz Jaskolski of the Faculty of Chemistry of A. Mickiewicz University in Poznan, Poland; and Iva Pichova of the Academy of Sciences of the Czech Republic, Prague.
The project was also supported by the UW Center for Game Science, the U.S. Defense Advanced Research Projects Agency (DARPA), the Howard Hughes Medical Institute, and the Microsoft Corporation.
-NSF-
-
Foldit players have control over which parts of proteins they'd like to move around.
Credit and Larger Version -
A Foldit introductory level tutorial for hydrogen-bonding; the player needs to move strands.
Credit and Larger Version -
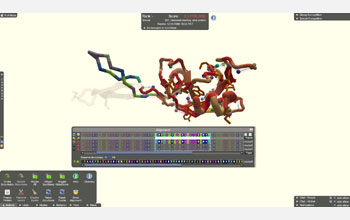
A screenshot of an active Foldit puzzle. The leaderboard shows the top score of each player/team.
Credit and Larger Version -
Foldit players use a variety of tools to interactively reshape various regions of proteins.
Credit and Larger Version -
Foldit "freezing:" a large red void in a protein's center shows empty space.
Credit and Larger Version
Media Contacts
Cheryl Dybas, NSF, (703) 292-7734, email: cdybas@nsf.gov
Leila Gray, University of Washington, (206) 685-0381, email: leilag@u.washington.edu
The U.S. National Science Foundation propels the nation forward by advancing fundamental research in all fields of science and engineering. NSF supports research and people by providing facilities, instruments and funding to support their ingenuity and sustain the U.S. as a global leader in research and innovation. With a fiscal year 2023 budget of $9.5 billion, NSF funds reach all 50 states through grants to nearly 2,000 colleges, universities and institutions. Each year, NSF receives more than 40,000 competitive proposals and makes about 11,000 new awards. Those awards include support for cooperative research with industry, Arctic and Antarctic research and operations, and U.S. participation in international scientific efforts.
Connect with us online
NSF website: nsf.gov
NSF News: nsf.gov/news
For News Media: nsf.gov/news/newsroom
Statistics: nsf.gov/statistics/
Awards database: nsf.gov/awardsearch/
Follow us on social
Twitter: twitter.com/NSF
Facebook: facebook.com/US.NSF
Instagram: instagram.com/nsfgov