News Release 07-139
Scientists Sequence Genome of Soil-Dwelling Green Alga
Results have implications for understanding early evolutionary events

Biologists have sequenced much of the genome of Chlamydomonas reinhardtii, a green alga.
October 11, 2007
This material is available primarily for archival purposes. Telephone numbers or other contact information may be out of date; please see current contact information at media contacts.
More than 100 scientists have reported an analysis of the genome of the green alga Chlamydomonas reinhardtii in the October 12 issue of the journal Science.
From a plethora of data on this one-celled soil-dweller, the researchers have found clues to the evolution of plants and animals, especially to the evolution of photosynthesis and of flagella, slender projections from a cell that propel it.
Chlamydomonas has long been a darling of the research lab. Its relative adaptability and short generation time (about 5 hours) have made it an important model organism for biological research.
Several years ago, scientists knew less than 2 percent of the sequence of the Chlamydomonas genome. Scientists now know 95 percent of the sequence of its genome.
Chlamydomonas's estimated 15,000 genes contain a wealth of information about the ancestry of plants and animals.
"Looking at the Chlamydomonas genome is like being in a candy store of thought," said Arthur Grossman, a plant biologist at the Carnegie Institution of Washington and a co-author of the Science paper. "From these analyses we're learning about the progenitor organism of animals and plants, the evolution and function of flagella, and how the different proteins in these structures relate to human diseases.
"We've also been able to identify classes of genes with unknown functions that are likely critical for photosynthesis."
The National Science Foundation (NSF) funded research performed by Grossman and collaborators; the U.S. Department of Energy's Joint Genome Institute supported the genome sequencing.
"Using comparative genomics to understand evolutionary relationships, especially those of the earliest ancestors of living organisms, is fundamental, exciting research," said James Collins, NSF assistant director for biological sciences. "The genome-enabled investigation of how an organism adapts to its environment, and how it communicates and competes with other organisms in the same community, impacts our understanding of basic biological principles. An understanding of those basic principles ultimately may be applied in a wide array of areas in biotechnology."
The scientists explored the evolutionary history of Chlamydomonas and its relationship to other organisms by comparing its genes with those of species from across the biological spectrum.
They found that Chlamydomonas shares 35 percent of its genes with both flowering plants and humans, an additional 10 percent with humans but not flowering plants, and 27 percent with flowering plants but not humans.
Chlamydomonas performs photosynthesis like plants--using the energy of sunlight, it converts carbon dioxide and water into oxygen and sugars. Indeed, much of what is known about photosynthesis was learned from Chlamydomonas.
The scientists identified protein families that are shared by Chlamydomonas, flowering plants, and other algae, but are not present in non-photosynthetic organisms. This enabled the researchers to recognize widely conserved photosynthesis-related proteins.
New proteins were identified in Chlamydomonas that are likely associated with flagella or the very similar cilia--thin organelles found in cells that provide locomotion and allow cells to sense their environment. Differentiated proteins that are critical for movement were also found in the alga, in addition to those associated with sensory functions.
Cilia are required for brain function and metabolism in animals. Humans have cilia in the brains, lungs and kidneys.
Loss of cilia leads to serious diseases, although scientists do not yet know why, said biochemist Sabeeha Merchant of UCLA. Chlamydomonas is ideal for discovering how cilia work, she said, and what goes wrong when they don't function properly.
-NSF-
-
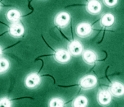
Chlamydomonas reinhardtii is a one-celled alga that shares genes with plants and animals.
Credit and Larger Version
Media Contacts
Cheryl Dybas, NSF, (703) 292-7734, email: cdybas@nsf.gov
Stuart Wolpert, UCLA, (310) 206-0511, email: swolpert@support.ucla.edu
Program Contacts
Susan Porter Ridley, NSF, (703) 292-8440, email: sridley@nsf.gov
Related Websites
NSF Directorate for Biological Sciences: http://www.nsf.gov/dir/index.jsp?org=BIO
The U.S. National Science Foundation propels the nation forward by advancing fundamental research in all fields of science and engineering. NSF supports research and people by providing facilities, instruments and funding to support their ingenuity and sustain the U.S. as a global leader in research and innovation. With a fiscal year 2023 budget of $9.5 billion, NSF funds reach all 50 states through grants to nearly 2,000 colleges, universities and institutions. Each year, NSF receives more than 40,000 competitive proposals and makes about 11,000 new awards. Those awards include support for cooperative research with industry, Arctic and Antarctic research and operations, and U.S. participation in international scientific efforts.
Connect with us online
NSF website: nsf.gov
NSF News: nsf.gov/news
For News Media: nsf.gov/news/newsroom
Statistics: nsf.gov/statistics/
Awards database: nsf.gov/awardsearch/
Follow us on social
Twitter: twitter.com/NSF
Facebook: facebook.com/US.NSF
Instagram: instagram.com/nsfgov