Multimedia Gallery
Improving Biomass Conversion
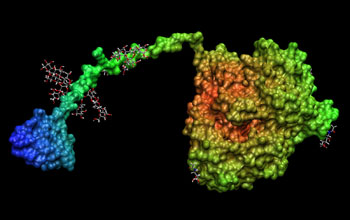
Improving biomass conversion to ethanol for renewable energy. To improve the conversion of biomass into ethanol, researchers from the National Renewable Energy Laboratory (NREL) simulated the action of the enzyme cellulase on cellulose using the CHARMM (Chemistry at HARvard Molecular Mechanics) community code. The binding domain (blue) is on the left, the glycosylated linker is green, and the catalyst domain, on the left, is in orange and yellow.
This work was carried out at the San Diego Supercomputer Center, which is supported in part by the National Science Foundation. (Date of Image: Sept. 9, 2004)
More about this Image
The National Renewable Energy Laboratory (NREL) is striving to develop efficient large-scale conversion of biomass into ethanol to provide a clean-burning and renewable fuel source. This can have benefits from reducing dependence on fossil fuels and imported oil to protecting the climate. A key bottleneck in making this process economically viable is the slow breakdown of cellulose by the enzyme cellulase, and scientists want to understand this process at the molecular level so that they can target further research to speed up this important reaction.
To explore the intricate molecular dynamics of this process, the researchers have used the CHARMM (Chemistry at HARvard Molecular Mechanics) code, a versatile community code for simulating biological reactions. But the size of new simulations needed is so large--more than one million atoms--and the simulation times so long--more than 5,000 timed steps for the 10 or more nanosecond simulation--that they exceed the current capabilities of CHARMM.
Now researchers at the San Diego Supercomputer Center (SDSC) are working with colleagues at NREL, Cornell, the Scripps Research Institute and the Colorado School of Mines to enhance CHARMM so that the simulations can scale up to millions of atoms and run on hundreds of processors on today's largest supercomputers, including SDSC's DataStar and the TeraGrid. This will make it feasible to simulate this key reaction. The research is enabling the largest simulations ever of an important scientific problem that will yield economic and environmental benefits and, in addition, the improvements to the CHARMM code will continue to be available for the scientific community to use on a wide range of other problems.
Credit: James Matthews, Linghao Zhong and John Brady, Cornell University; Mike Himmel and Mark Nimlos, NREL; Tauna Rignall, Colorado School of Mines; Mike Crowley, The Scripps Research Institute. Work was funded by an NREL subcontract funded by DOE office of the
See other images like this on your iPhone or iPad download NSF Science Zone on the Apple App Store.
Special Restrictions: James Matthews, Linghao Zhong and John Brady, Cornell University; Mike Himmel and Mark Nimlos, NREL; Tauna Rignall, Colorado School of Mines; Mike Crowley, Scripps Institute. Work funded by NREL subcontract funded by DOE Office of the Biomass Program.
Images and other media in the National Science Foundation Multimedia Gallery are available for use in print and electronic material by NSF employees, members of the media, university staff, teachers and the general public. All media in the gallery are intended for personal, educational and nonprofit/non-commercial use only.
Images credited to the National Science Foundation, a federal agency, are in the public domain. The images were created by employees of the United States Government as part of their official duties or prepared by contractors as "works for hire" for NSF. You may freely use NSF-credited images and, at your discretion, credit NSF with a "Courtesy: National Science Foundation" notation.
Additional information about general usage can be found in Conditions.
Also Available:
Download the high-resolution JPG version of the image. (429 KB)
Use your mouse to right-click (Mac users may need to Ctrl-click) the link above and choose the option that will save the file or target to your computer.